Overview
C-It-Loci is a tool that contains four types of data: "Conserved Regions", “Genes", “Transcripts", and "Expressed Transcripts".
Three of these are searchable/browseable via their corresponding links. Each row within these pages links to a page
containing RNA-seq data from a variety of tissues with other useful data.
Each data type can be accessed by clicking the corresponding link on the navigation bar at the top of the
page. This will direct to the page containing search form and a table of entries.
See Browsing and Searching below for directions on navigating and searching C-It-Loci.
CGP: “
C-It-Loci
Genome
Position”, which is a region in which pairs of adjacent homologous
protein-coding genes shared between one or more species.
UCNEbase: regions defined by the database for ultra-conserved non-coding elements
(UCNEs) called “UCNEbase” (
http://ccg.vital-it.ch/UCNEbase/).
For details about UCNEbase, please see: Dimitrieva S, Bucher P (2013) UCNEbase--a database of
ultraconserved non-coding elements and genomic regulatory blocks.
Nucleic Acids Res 41(Database issue):D101-9.
VISTA: loci that are +/-300kb from the regions defined by the VISTA Enhancer Browser
(http://enhancer.lbl.gov). For details about the VISTA Enhancer Browser, please
see: Visel A, Minovitsky S, Dubchak I, Pennacchio LA (2007) VISTA Enhancer
Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35(Database issue):D88-92.
It is advised to use C-It-Loci's advanced search. However, in many cases, simplified search offers a useful and
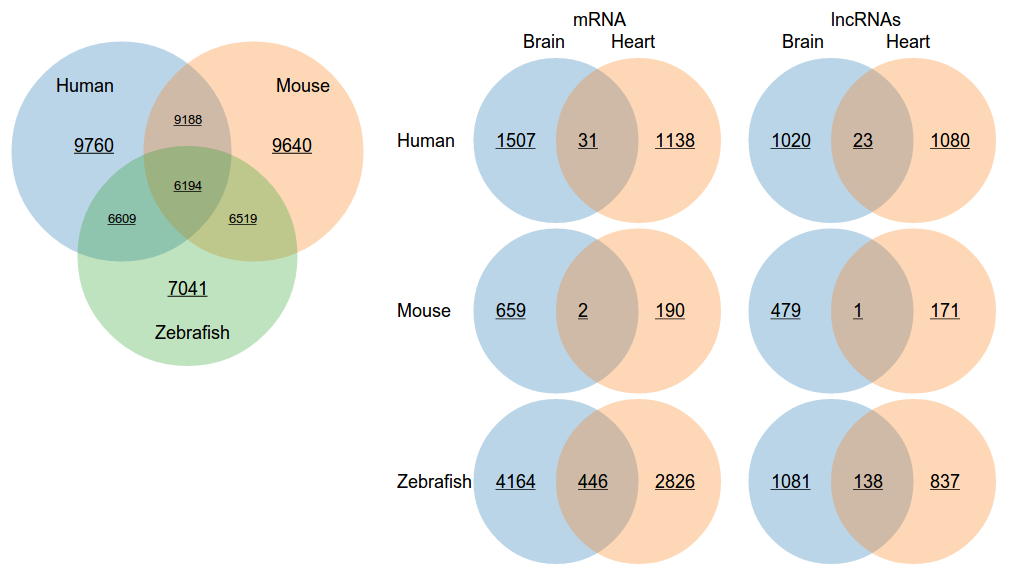
simple graphical method to search and to compare data. Upon submission, quick search will return Venn diagrams
displaying the numbers of conserved regions, protein-coding transcripts, and lncRNA transcripts.
- Select tissue 1 (Required) - Specifies the primary tissue for the search, which is shown as the blue circles of the Venn diagrams on the left.
- Select expression 1 (Required) - Determines the type of expression shown by the Venn diagrams.
- Select tissue 2 (Optional) - Allows for comparing a secondary tissue to the primary tissue, which is shown as orange circles on the right of Venn diagrams.
- Select expression 2 (Optional) - Determines the type of expression shown by the Venn diagrams.
- GO - Clinking this button will submit the query.
To preform a quick search, the top selection boxes for a tissue and expression type must contain a
tissue and expression value. The second row of tissues and expression types is optional, however, both
must be set for the form to be valid. Note: the 'GO' button will be disabled if the form is not valid.
The advance search allows for greater control of search results by allowing queries based on various types of
data included in C-It-Loci. The advance search uses a query tag combined with a search string to specify the type
of data to return. Additionally, the output of these queries can be combined together using the boolean operators: “and", “or", “not", and “()”.
Note, if a query contains a space, then quotation marks should be added around the full string.
If no tag is present in the query, it will be treated as an accession number or gene name.
Again, if any spaces are included within the string, the entire string will be treated as multiple
queries unless surrounded by quotation marks. See the table below for the types of queries:
| Expression |
Example |
Description |
| No tag. |
CIL_000001 |
When no tag is specified the query is treated as an accession or gene name |
| SPECIFIC:(tissue) |
SPECIFIC:Heart |
Returns transcripts detected in only one type of tissue. |
| ENRICHED:(tissue) |
ENRICHED:Heart |
Returns transcripts which have increased expression in the given tissue. |
| EXPRESSED:(tissue) |
EXPRESSED:Heart |
Returns transcripts expressed in the given tissue type. |
| FPKMGT:(FPKM) or FPKMGT:(FPKM):(tissue) |
FPKMGT:5 or FPKMGT:5:Heart |
Returns transcripts with expression greater than the given FPKM. If a tissue is passed this will
return transcripts with expression greater than the given FPKM in the given tissue.
|
| FPKMLT:(FPKM) or FPKMLT:(FPKM):(tissue) |
FPKMLT:5 or FPKMLT:5:Heart |
Returns transcripts with expression less than the given FPKM. If a tissue is passed this will
return transcripts with expression less than the given FPKM in the given tissue.
|
| POS:(tax_id):(chromosome):(start)-(end) |
POS:9606:1:914880-969309 |
Returns regions, genes, or transcripts falling withing the given position. |
| BIOTYPE:(biotype) |
BIOTYPE:lincRNA |
Returns regions, genes, or transcripts which contain a transcript annotated as the given biotype. |
| REGIONTYPE:(CGP|UCNEbase|VISTA) |
REGIONTYPE:UCNEbase |
Returns regions which contain the given region type. If searching for genes or transcripts,
genes and transcripts contained within the given region type will be returned.
|
| TAXID:(7955|9606|10090) |
TAXID:7955 |
Returns regions, genes, or transcripts associated with the given taxonomy.
Taxonomy IDs (TAXID) are: 7955 (zebrafish), 9606 (human), and 10090 (mouse). |
| GO:(GO term accession) |
GO:0005515 |
Returns regions, genes, or transcripts which contain an annotation of the given Gene Ontology (GO) term. |
| Boolean Operators |
|---|
| and |
TAXID:7655 and TAXID:10090 |
Returns accessions existing between two query sets. |
| or |
BIOTYPE:lincRNA or BIOTYPE:antisense |
Returns accessions existing in either of two query sets. |
| not |
not TAXID:9606 |
Returns accessions not in the query. |
| () | BIOTYPE:lincRNA and
(TAXID:7955 and TAXID:10090 and not TAXID:9606) or
(TAXID:9606 and TAXID:7955 and not TAXID:10090) |
Increases the precedence of a query. |
| * |
gata* |
Specifies where to use wildcards in query. Note: wildcards are only supported for tagless searches. |
The large size of C-It-Loci necessitates breaking queries into manageable sizes. This is accomplished via the
browsing view. The browsing view breaks queries into pages of a given number of rows and allows navigation between
pages via the “Previous” and “Next” buttons. Navigation to a desire page is also possible by setting the page
number field to the desired page.

- The number of rows to show per page.
- Displays the number of the current page. When changed this will redirect to the new page number
- Move to the previous page.
- Move to the next page.
Record pages contain detailed information about a data type contained within C-It-Loci.
This can include annotation data, genomic coordinates, expression data, homologs, and GO
terms depending on the data type.
Each record page will contain at least one genome view. C-It-Loci uses the Biodalliance genome browser.
For details about this browser, please refer to
http://www.biodalliance.org/
There are two ways to view expression data: a graphical view and a text view. Users can toggle between the two
views using the provided buttons. The text view is a simple table comparing various transcript expressions
versus tissues with numerical values representing the average FPKM values followed by the standard
deviations with ± symbol. The graphical view scales the FPKM values using the equation
y = 100.0 - (log(x)+12)*((3*x)/(x+1)+1), where y is the lightness value in the HSLA color.
Downloads
- bed file with conserved regions - Download
- All expression data:
| Study Accession | Sample Accession | Run Accession | tax_id | Instrument Model | Library Layout | Age | Tissue | ENSEMBL Transcripts Detected |
|---|
| ERP000447 | SAMEA782568 | ERR023144 | 7955 | Genome Analyzer II | PAIRED | Adult | Brain | 16936 |
| ERP000447 | SAMEA782570 | ERR023145 | 7955 | Genome Analyzer II | PAIRED | Adult | Heart | 10856 |
| ERP000447 | SAMEA782572 | ERR023146 | 7955 | Genome Analyzer II | PAIRED | Adult | Kidney | 13547 |
| ERP000447 | SAMEA782568 | ERR023147 | 7955 | Genome Analyzer II | PAIRED | Adult | Brain | 16602 |
| ERP000447 | SAMEA782572 | ERR023149 | 7955 | Genome Analyzer II | PAIRED | Adult | Kidney | 13198 |
| ERP000447 | SAMEA782570 | ERR023150 | 7955 | Genome Analyzer II | PAIRED | Adult | Heart | 11439 |
| ERP000447 | SAMEA782568 | ERR035545 | 7955 | Genome Analyzer II | PAIRED | Adult | Brain | 23167 |
| ERP000447 | SAMEA782570 | ERR035546 | 7955 | Genome Analyzer II | PAIRED | Adult | Heart | 23476 |
| ERP000447 | SAMEA782572 | ERR035547 | 7955 | Genome Analyzer II | PAIRED | Adult | Kidney | 24427 |
| SRP012040 | SAMN00849374 | SRR453077 | 10090 | Genome Analyzer II | PAIRED | Adult | Ovary | 23673 |
| SRP012040 | SAMN00849374 | SRR453078 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 23249 |
| SRP012040 | SAMN00849374 | SRR453079 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 21858 |
| SRP012040 | SAMN00849374 | SRR453080 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 23178 |
| SRP012040 | SAMN00849374 | SRR453081 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 23421 |
| SRP012040 | SAMN00849374 | SRR453082 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 22867 |
| SRP012040 | SAMN00849374 | SRR453083 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 22952 |
| SRP012040 | SAMN00849374 | SRR453084 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 22586 |
| SRP012040 | SAMN00849374 | SRR453085 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 23071 |
| SRP012040 | SAMN00849374 | SRR453086 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Ovary | 23121 |
| SRP012040 | SAMN00849375 | SRR453087 | 10090 | Genome Analyzer II | PAIRED | Adult | Breast | 21881 |
| SRP012040 | SAMN00849375 | SRR453088 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Breast | 24261 |
| SRP012040 | SAMN00849375 | SRR453089 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Breast | 24505 |
| SRP012040 | SAMN00849375 | SRR453090 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Breast | 22846 |
| SRP012040 | SAMN00849375 | SRR453091 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Breast | 22780 |
| SRP012040 | SAMN00849375 | SRR453092 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Breast | 23724 |
| SRP012040 | SAMN00849379 | SRR453116 | 10090 | Genome Analyzer II | PAIRED | Adult | Adrenal | 21591 |
| SRP012040 | SAMN00849379 | SRR453117 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adrenal | 21496 |
| SRP012040 | SAMN00849379 | SRR453118 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adrenal | 21494 |
| SRP012040 | SAMN00849379 | SRR453119 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adrenal | 21648 |
| SRP012040 | SAMN00849379 | SRR453120 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adrenal | 21854 |
| SRP012040 | SAMN00849379 | SRR453121 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adrenal | 21502 |
| SRP012040 | SAMN00849382 | SRR453130 | 10090 | Genome Analyzer II | PAIRED | Adult | Adipose | 24279 |
| SRP012040 | SAMN00849382 | SRR453131 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adipose | 24709 |
| SRP012040 | SAMN00849382 | SRR453132 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adipose | 23737 |
| SRP012040 | SAMN00849382 | SRR453133 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Adipose | 23366 |
| SRP012040 | SAMN00849384 | SRR453140 | 10090 | Genome Analyzer II | PAIRED | Adult | Testis | 27217 |
| SRP012040 | SAMN00849384 | SRR453141 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Testis | 27203 |
| SRP012040 | SAMN00849384 | SRR453142 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Testis | 27106 |
| SRP012040 | SAMN00849384 | SRR453143 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Testis | 26915 |
| SRP012040 | SAMN00849385 | SRR453144 | 10090 | Genome Analyzer II | PAIRED | Adult | Kidney | 24230 |
| SRP012040 | SAMN00849385 | SRR453145 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Kidney | 24150 |
| SRP012040 | SAMN00849385 | SRR453146 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Kidney | 22884 |
| SRP012040 | SAMN00849385 | SRR453147 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Kidney | 24114 |
| SRP012040 | SAMN00849385 | SRR453148 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Kidney | 23974 |
| SRP012040 | SAMN00849385 | SRR453149 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Kidney | 23895 |
| SRP012040 | SAMN00849386 | SRR453150 | 10090 | Genome Analyzer II | PAIRED | Adult | Liver | 20467 |
| SRP012040 | SAMN00849386 | SRR453151 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Liver | 19505 |
| SRP012040 | SAMN00849386 | SRR453152 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Liver | 20444 |
| SRP012040 | SAMN00849386 | SRR453153 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Liver | 20987 |
| SRP012040 | SAMN00849386 | SRR453154 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Liver | 20716 |
| SRP012040 | SAMN00849386 | SRR453155 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Liver | 21090 |
| SRP012040 | SAMN00849387 | SRR453156 | 10090 | Genome Analyzer II | PAIRED | Adult | Lung | 24819 |
| SRP012040 | SAMN00849387 | SRR453157 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Lung | 24701 |
| SRP012040 | SAMN00849387 | SRR453158 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Lung | 24290 |
| SRP012040 | SAMN00849387 | SRR453159 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Lung | 24146 |
| SRP012040 | SAMN00849389 | SRR453166 | 10090 | Genome Analyzer II | PAIRED | Adult | Colon | 22852 |
| SRP012040 | SAMN00849389 | SRR453167 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Colon | 22767 |
| SRP012040 | SAMN00849389 | SRR453168 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Colon | 22144 |
| SRP012040 | SAMN00849389 | SRR453169 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Colon | 23415 |
| SRP012040 | SAMN00849389 | SRR453170 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Colon | 23206 |
| SRP012040 | SAMN00849389 | SRR453171 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Colon | 23291 |
| SRP012040 | SAMN00849390 | SRR453172 | 10090 | Genome Analyzer II | PAIRED | Adult | Heart | 22709 |
| SRP012040 | SAMN00849390 | SRR453173 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Heart | 22627 |
| SRP012040 | SAMN00849390 | SRR453174 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Heart | 23192 |
| SRP012040 | SAMN00849390 | SRR453175 | 10090 | Genome Analyzer II | PAIRED | adult-8wks | Heart | 23164 |
| SRP012040 | SAMN01164133 | SRR567482 | 10090 | HiSeq 2000 | PAIRED | Adult | Bladder | 26140 |
| SRP012040 | SAMN01164133 | SRR567483 | 10090 | HiSeq 2000 | PAIRED | adult-8wks | Bladder | 25423 |
| SRP012040 | SAMN01164134 | SRR567484 | 10090 | HiSeq 2000 | PAIRED | Adult | Placenta | 26598 |
| SRP012040 | SAMN01164134 | SRR567485 | 10090 | HiSeq 2000 | PAIRED | adult-8wks | Placenta | 27056 |
| SRP012040 | SAMN01164135 | SRR567486 | 10090 | HiSeq 2000 | PAIRED | Embyro | Liver | 24957 |
| SRP012040 | SAMN01164135 | SRR567487 | 10090 | HiSeq 2000 | PAIRED | E18 | Liver | 24380 |
| SRP012040 | SAMN01164140 | SRR567496 | 10090 | HiSeq 2000 | PAIRED | Embyro | Liver | 25628 |
| SRP012040 | SAMN01164140 | SRR567497 | 10090 | HiSeq 2000 | PAIRED | E14.5 | Liver | 22850 |
| SRP012040 | SAMN01164141 | SRR567498 | 10090 | HiSeq 2000 | PAIRED | Embyro | Brain | 26181 |
| SRP012040 | SAMN01164141 | SRR567499 | 10090 | HiSeq 2000 | PAIRED | E14.5 | Brain | 27856 |
| SRP012040 | SAMN01164143 | SRR567502 | 10090 | HiSeq 2000 | PAIRED | Embyro | Liver | 25365 |
| SRP012040 | SAMN01164143 | SRR567503 | 10090 | HiSeq 2000 | PAIRED | E14 | Liver | 24369 |
| SRP017959 | SAMN01886753 | SRR649362 | 9606 | Illumina Genome Iix | SINGLE | Adult | Testis | 44064 |
| SRP017959 | SAMN01886754 | SRR649363 | 9606 | Illumina Genome Iix | SINGLE | Embyro | Placenta | 32522 |
| SRP017959 | SAMN01886755 | SRR649364 | 9606 | Illumina Genome Iix | SINGLE | Adult | Ovary | 34352 |
| SRP017959 | SAMN01886761 | SRR649372 | 10090 | Illumina Genome Iix | SINGLE | Adult | Ovary | 27504 |
| SRP017959 | SAMN01886762 | SRR649373 | 10090 | Illumina Genome Iix | SINGLE | Embyro | Placenta | 24487 |
| SRP017959 | SAMN01886763 | SRR649374 | 10090 | Illumina Genome Iix | SINGLE | Embyro | Placenta | 24700 |
| SRP017959 | SAMN01886764 | SRR649375 | 10090 | Illumina Genome Iix | SINGLE | Adult | Testis | 27413 |
| SRP019807 | SAMN01984805 | SRR787270 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Bladder | 25830 |
| SRP019807 | SAMN01984806 | SRR787271 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Brain | 28033 |
| SRP019807 | SAMN01984807 | SRR787272 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Breast | 27519 |
| SRP019807 | SAMN01984808 | SRR787273 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Colon | 26932 |
| SRP019807 | SAMN01984809 | SRR787274 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Heart | 26587 |
| SRP019807 | SAMN01984810 | SRR787275 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Kidney | 22583 |
| SRP019807 | SAMN01984811 | SRR787276 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Liver | 27075 |
| SRP019807 | SAMN01984812 | SRR787277 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Lung | 26862 |
| SRP019807 | SAMN01984813 | SRR787278 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Muscle | 19319 |
| SRP019807 | SAMN01984814 | SRR787279 | 9606 | Illumina HiSeq 2000 | PAIRED | NA | Ovary | 26467 |
| SRP024369 | SAMN02951650 | SRR891495 | 7955 | Illumina Genome Analyzer IIx | PAIRED | Adult | Heart | 25049 |
| SRP024369 | SAMN02951649 | SRR891504 | 7955 | Illumina Genome Analyzer IIx | PAIRED | Adult | Liver | 22481 |
| SRP024369 | SAMN02951652 | SRR891510 | 7955 | Illumina Genome Analyzer IIx | PAIRED | Adult | Muscle | 24932 |
| SRP024369 | SAMN02951648 | SRR891511 | 7955 | Illumina Genome Analyzer IIx | PAIRED | Adult | Brain | 24478 |
| SRP017959 | SAMN02265389 | SRR943340 | 9606 | Illumina HiSeq 2000 | SINGLE | Embyro | Placenta | 26721 |
| SRP017959 | SAMN02265388 | SRR943341 | 9606 | Illumina HiSeq 2000 | SINGLE | NA | Ovary | 31273 |
| SRP017959 | SAMN02265387 | SRR943342 | 10090 | Illumina HiSeq 2000 | SINGLE | Adult | Ovary | 24365 |
| SRP017959 | SAMN02265390 | SRR943343 | 10090 | Illumina HiSeq 2000 | SINGLE | Adult | Ovary | 24233 |
| SRP017959 | SAMN02265391 | SRR943344 | 10090 | Illumina HiSeq 2000 | SINGLE | Embyro | Placenta | 24454 |
| SRP017959 | SAMN02265393 | SRR943345 | 10090 | Illumina HiSeq 2000 | SINGLE | Embyro | Placenta | 24388 |
| SRP017959 | SAMN02265400 | SRR943355 | 10090 | Illumina HiSeq 2000 | SINGLE | Embyro | Placenta | 27713 |
| SRP017959 | SAMN01886760 | SRR649371 | 10090 | Illumina Genome Analyzer Iix | SINGLE | Adult | Brain | 26157 |